
Couples who have PGD will undergo an in vitro fertilization (IVF) cycle to create embryos. Genetic analysis will then be performed on cells from each embryo prior to transfer into the woman’s uterus. To analyze an embryo, we biopsy the embryo around the third day of its development when the embryo has approximately 6-8 cells. One or two cells are taken from the embryo. The embryo is incubated until testing is complete.
The biopsied cells are analyzed using a technique called fluorescence in-situ hybridization or FISH. This technique uses probes, small pieces of DNA that are a match for the chromosomes we want to analyze, to count the chromosomes present. FISH is the most commonly applied method to determine the chromosomal constitution of an embryo. In contrast to karyotyping, it can be used on interphase chromosomes, so that it can be used on PBs and blastomeres. The biopsied PBs or blastomeres are first fixed to a microscope slide and then the cellular material is digested away leaving the nucleus, which contains the DNA, in a spread out and decondensed form. These cells are then hybridised with DNA probes, labeled with different fluorochromes. Each of these probes are specific for part of a chromosome; they will only attach to their exact DNA match on a particular chromosome. Excess probe is washed off, and the cell is examined under the fluorescent microscope. We then count the number of chromosomes of each type (color) there are in that cell. The geneticist therefore can distinguish normal cells from cells with aneuploidy.
Currently, a large panel of probes are available for different segments of all chromosomes, but the limited number of different fluorochromes confines the number of signals that can be analysed simultaneously. The type and number of probes that are used on a sample depends on the indication. For sex determination (used for instance when a PCR protocol for a given X-linked disorder is not available), probes for the X and Y chromosomes are applied along with probes for one or more of the autosomes as an internal FISH control. More probes can be added to check for aneuploidies, particularly those that could give raise to a viable pregnancy (such as a trisomy 21). Genoma's PGD laboratory uses FISH probes for the chromosomes most commonly found in abnormalities: chromosomes X, Y, 13, 14, 15, 16, 18, 21 and 22. This panel of probes has the potential of detecting 70% of the aneuploidies found in spontaneous abortions.
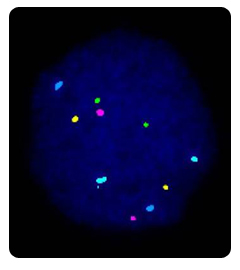
In order to be able to analyse more chromosomes on the same sample, up to three consecutive rounds of FISH can be carried out. In the case of chromosome rearrangements, specific combinations of probes have to be chosen that flank the region of interest.
Testing of the cells destroys them because they must be glued to a glass slide and repeatedly heated and cooled. As such, one cannot use them for another purpose or return them to the embryo. The slides are kept for future reference. This analysis causes no extra inconvenience to the patient as it is accomplished in one day.
The FISH technique is considered to have an error rate between 5 and 10%. The main problem of the use of FISH to study the chromosomal constitution of embryos is the elevated mosaicism rate observed at the human preimplantation stage. Sandalinas and collaborators found that up to 70% of the embryos they studied by FISH were mosaic for some kind of chromosomal abnormality (Sandalinas et al., 2001). Li and co-workers (2005) found that 40% of the embryos diagnosed as aneuploid on day 3 turned out to have a euploid inner cell mass at day 6. Staessen and collaborators found that 17.5% of the embryos diagnosed as abnormal during PGS, and subjected to post-PGD reanalysis, were found to also contain normal cells, and 8.4% were found grossly normal (Staessen et al., 2004). As a consequence, it has been questioned whether the one or two cells studied from an embryo are actually representative of the complete embryo, and whether viable embryos are not being discarded due to the limitations of the technique.