The diagnostic protocol involves a fluorescent multiplex PCR analysis of highly polymorphic STR markers, linked to the disease causing genes, to identify the haplotype associated with the maternal mutation. In fact, any informative polymorphism, which lies in close proximity to the disease causing gene, can be used as a tool to ascertain the presence or absence of the mutation, without its direct detection, just evaluating the inheritance of the haplotypes, obtained by segregation analysis of the STR alleles (Figure 2).
Because 1PB is the mirror image of the oocyte, its genotype is indirectly derived from the opposite diagnosis of the 1PB.The presence of the haplotype associated with the mutated allele in 1PB indicates that both copies of the mutated gene have been extruded and the oocyte can be predicted to be free of mutation. On the contrary, if 1PB analysis reveals the presence of the haplotype associated with the normal allele, the oocyte presumptively contains the mutant gene. If crossing over does occur, the 1PB will contain both the normal and the mutated haplotype, showing a heterozygous genotype for the STR markers investigated (Figure 1).
The implementation of an indirect diagnosis, by haplotype analysis, precludes the need of detecting the specific causative mutation, and therefore provides a unique protocol applicable to most of the patients at risks of transmitting a specific gene defect. This approach can be carried out for any mutations combination, without the need to develop a specific diagnostic experimental design for each couple. As a consequence, a substantial shortening of the preclinical work-up time necessary for each case can be achieved.
A panel of 6 closely linked highly polymorphic STR markers is studied, to ensure a sufficient informativity in all cases and also to avoid misdiagnosis in recombinant oocytes due to possible allele drop-out (ADO) occurrences. The co-amplification of several polymorphic markers reduces the risk of amplification failure and increases the assay accuracy by allowing the detection of potential ADO occurring in multiple markers, which would lead to diagnose a recombinant heterozygous oocyte as hemizygous, thus reducing substantially the risk of misdiagnosis. In fact, in such a case, misdiagnosis is only possible in the very unlikely hypothesis that ADO of the wild-type allele occurs in all amplified markers.
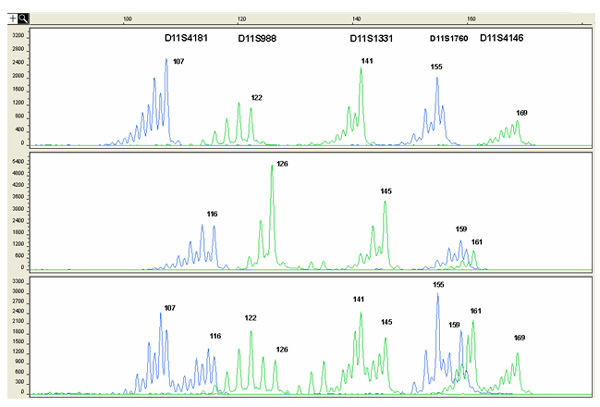 Figure 2. Example of electropherograms showing the fluorescent PCR products obtained from 1PB after multiplex amplification of 5 informative STR markers flanking the HBB gene. The x-axis shows length of PCR products in bp and the y-axis shows the fluorescence intensity in Relative Fluorescence Units (RFU). On top of the electropherogram the marker name is located above the corresponding alleles (peaks). Numbers next to each peak represent the size of the allele (in bp). The upper lane shows the haplotype associated with the maternal mutation, therefore the corresponding oocytes is diagnosed as mutation-free. The haplotype associated with the normal allele is shown in the middle lane, thus the corresponding oocyte contains the mutant gene. The lower lane shows the profile of a carrier female, presenting both the mutant and the normal haplotype. The maternal haplotype was established by family studies.
Figure 2. Example of electropherograms showing the fluorescent PCR products obtained from 1PB after multiplex amplification of 5 informative STR markers flanking the HBB gene. The x-axis shows length of PCR products in bp and the y-axis shows the fluorescence intensity in Relative Fluorescence Units (RFU). On top of the electropherogram the marker name is located above the corresponding alleles (peaks). Numbers next to each peak represent the size of the allele (in bp). The upper lane shows the haplotype associated with the maternal mutation, therefore the corresponding oocytes is diagnosed as mutation-free. The haplotype associated with the normal allele is shown in the middle lane, thus the corresponding oocyte contains the mutant gene. The lower lane shows the profile of a carrier female, presenting both the mutant and the normal haplotype. The maternal haplotype was established by family studies.