The biopsied cells are analyzed using a technique called the polymerase chain reaction (PCR). PCR is an in vitro simplified reproduction of the in vivo process of DNA replication. Taking advantage from the chemical properties of DNA and the availability of thermostable DNA polymerases, PCR allows for the enrichment of a DNA sample for a certain sequence.
PCR provides the possibility to obtain a large quantity of copies of a particular stretch of the genome, making further analysis possible.
It is a highly sensitive and specific technology, which makes it suitable for all kinds of genetic diagnosis, including PGD.
Each cell contains a minute amount of DNA (a single copy). PCR is used to amplify the DNA to a detectable level. Once amplification has been accomplished scientists can use a variety of techniques to screen an individual gene for abnormalities.
This procedure is known as Mutation Analysis.
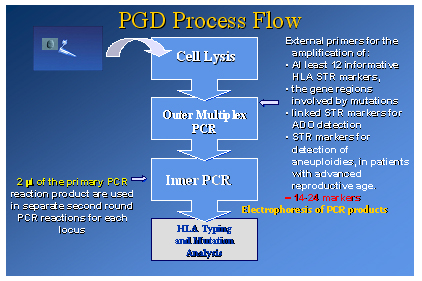 Multiplex PCR
Multiplex PCR
Before proceeding to multiplex PCR, cells were lysed by incubation at 65°C for 10 min. The alkaline lysis buffer was then neutralized by the addition of 5 ml of neutralization buffer.
A nested multiplex PCR assay is used to co-amplify all the selected loci.
The first round PCR containes the external primers for the amplification of the informative HLA STR markers selected during the preclinical work-up of each PGD case, the gene regions involved by mutations, STR markers linked to these regions for ADO detection and STR markers used for detection of aneuploidies in patients of advanced reproductive age. The first round multiplex PCR is followed by separate second round PCR reactions for each locus, using 2 ml of the primary PCR products.
The HLA complex
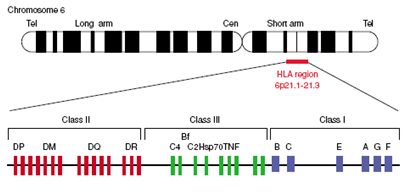
Peptides encoded within the major histocompatibility complex (MHC) have the ability to provoke vigorous rejection of grafts in transplantations (Roitt, 1994). The complex, also referred to as human leukocyte antigen (HLA) region, can be divided in 3 regions: class I, class II and class III (Figure).
Class I and II molecules are membrane-bound proteins which are recognized by T-cells; class I molecules signal for cytotoxic T-cells, while class II signal T-helpers for B-cells and macrophages. Class III molecules include complement components, heat sock proteins and tumor necrosis factors. The recognition of the class I and II molecules by T-cells mediates the graft rejection response in transplantations. For this reason, histocompatibility testing is mainly focused in these two regions. Class I HLA-A and HLA-B molecules are expressed abundantly and are highly polymorphic.
HLA-DP,-DQ and –DR are also polymorphic. Population studies have shown that matching of the above alleles is associated with lower risk of graft failure; moreover, clinical studies have demonstrated that the use of molecular methods for HLA typing improves the efficiency of haematopoietic cell transplantation (Petersdorf et al., 2003).
HLA haplotying
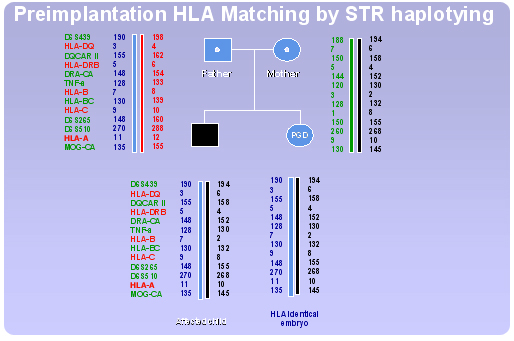
Technically, PGD for HLA typing is a difficult procedure due to the extreme polymorphism of the HLA region. Taking into account also the complexity of the region (presence of a large number of loci and alleles) the use of a direct HLA typing approach would require standardization of a PCR protocol specific for each family, presenting different HLA allele combinations, making it time consuming and unfeasible. The use of a preimplantation HLA matching protocol irrespective of the specific genotypes involved facilitates notably the procedure.
Our Centre has developed a new strategy based on the use of a flexible indirect HLA typing protocol applicable to a wide spectrum of possible HLA genotypes. The approach involves testing of single blastomeres by fluorescent multiplex polymerase chain reaction (PCR) analysis of polymorphic short tandem repeat (STR) markers, scattered throughout the HLA complex, obtaining a “fingerprint” of the entire HLA region.
By selecting a consistent number of STR markers evenly spaced throughout the HLA complex, an accurate mapping of the whole region can be achieved. Because genes in the HLA complex are tightly linked and usually inherited in block, profiles obtained from such markers in father, mother and affected child allow the determination of specific haplotypes. Thus, the HLA region can be indirectly typed by segregation analysis of the STR alleles and the HLA identity of the embryos with the affected sibling can be ascertained evaluating the inheritance of the matching haplotypes.
The use of microsatellite markers for this purpose is very useful, since they may provide information on identity over a greater distance within the HLA region compared to matching strictly for the classic HLA genes, making haplotyping more accurate in predicting compatibility.
The strategy presented here enables the selection of HLA-matched embryos can be performed for any genotype combination, without the need to develop a specific diagnostic experimental design for each couple, because the selected panel of STR markers have already been worked out and can be used for other patients. As a consequence, a substantial shortening of the preliminary phase can be achieved.
It is important to keep the time to develop a family-specific PCR protocol as short as possible; using STR markers enables to work quickly and safely. Obviously there is a positive impact on overall cost-effectiveness but there are also clinical arguments. Patients with congenital anemias can survive for many years. However, they will develop a number of time-dependent complications causing direct morbidity and mortality and reducing the success-rate of an eventual transplant procedure. For patients with congenital immune deficiencies, the continuous risk of potentially dangerous infections will decrease if the transplant is performed earlier. In urgent cases such as leukemia, the children are in remission but they can relapse suddenly and therefore there is no time to lose. Finally it is important to speed up the preclinical work to reduce the psychological stress the family is confronted with while waiting.
An example of preimplantation HLA matching procedure using STR haplotyping, in combination with PGD for b-thalassaemia, is shown in Figure 2. A total of 15 cumulus-oocyte complexes (COCs) were retrieved for this PGD cycle, 12 mature oocytes were inseminated by ICSI and 12 became fertilised. Three days later 12 embryos were selected for embryo biopsy on the basis of regular development and morphology. Two blastomeres were removed and analysed from each embryo. Eleven embryos yielded conclusive results, 8 of which resulted HLA non-identical (2 affected, 5 carrier, 1 normal). Only three embryos appeared to be both healthy and HLA identical and were therefore transferred, resulting in the birth of two twins HLA matched with the affected sibling. Stem cells collected from the umbilical cord blood of both twins were transplanted to the affected child, who is no longer blood transfusion dependent.
Figure 3 presents the results of preimplantation HLA matching performed in combination with PGD for WAS. A total of 15 regularly fertilized oocytes resulted after inseminating 19 oocytes. On the third day, 13 embryo were suitable for biopsy, in 6 of which only 1 cell was removed. From 7 embryos 2 blastomeres were collected. Eleven embryos produced conclusive results, 9 of which resulted HLA non-identical (2 affected, 1 carrier, 6 normal). Only 2 HLA-matched embryos were obtained, but only one (embryo 14) appeared also unaffected and was therefore transferred, resulting in birth of a carrier female, HLA matched with the affected sibling. Stem cell transplantation was performed in the affected child, resulting in a successful hematopoietic reconstruction.
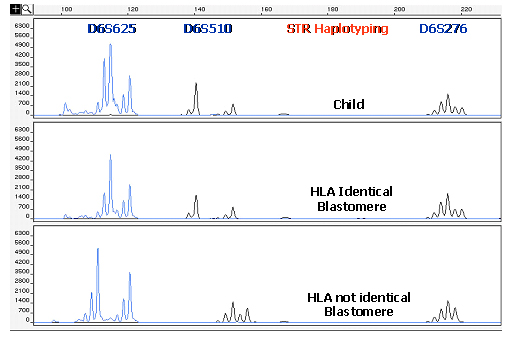
Another important advantage of using STR markers in preimplantation HLA matching is that the whole HLA complex can be covered and this allows the detection of recombination events between HLA genes. Recombination occurrences, if not detected, could strongly affect the accuracy of the HLA matching procedure.
The importance of detecting recombination within the HLA region is demonstrated in Figure 4, where are described the results of a cycle involving preimplantation HLA matching, without PGD of a causative gene, performed for a couple having a child affected by a sporadic form of DBA. In this case, embryos were genotyped using a panel of 14 different STR markers, evenly distributed along the whole HLA region. Recombination between flanking markers of the paternal or maternal haplotype was detected in 2 (embryos 1 and 6) of the 15 embryos tested. In one of them (embryo 1), a single recombination occurred in the maternal haplotype, between the alleles of the markers D6S105 and MIB. In the other embryo (embryo 6), initially appearing to be HLA matched with the affected sibling, a double recombination event was observed between markers D6S1683 and D6S265. This occurrence, detected by using a consistent number of STR markers able to determine a fine mapping of the whole HLA region, could lead to a HLA-genotyping misdiagnosis if not detected, and the embryo would be erroneously diagnosed as HLA identical. Hence, the reliability of the procedure is strongly correlated with the number of STR markers used for HLA haplotyping.
The combined use of a multiplex HLA STR marker system also allows detection of aneuploidies of chromosome 6. The relevance of aneuploidy testing for chromosome 6 is seen in Figure 4. One of the 15 embryos tested in this case appeared to have only one maternal chromosome 6 (embryos 5), and one (embryo 13) had an extra maternal chromosome, consistent with a diagnosis of monosomy 6 and trisomy 6, respectively, making them unacceptable for transfer.